Krishna Rajarathnam, PhD
Krishna Rajarathnam, PhD
Professor, Department of Biochemistry & Molecular Biology
Tel: (409) 772-2238
Fax: (409) 777-4738
E-mail: krrajara@utmb.edu
Campus Location: 5.142 Medical Research Bldg
Mail
Route: 1055
Research
Our lab is interested in understanding the molecular mechanisms by which chemokines activate GPCR receptors and bind sulfated glycosaminoglycans (GAGs) to orchestrate in vivo leukocyte recruitment to the infected tissue and activate leukocytes for microbial killing at the target site. Precise spatiotemporal control of these processes is essential to mount an effective defense. Impaired recruitment and/or impaired activation will result in incomplete resolution, whereas uncontrolled recruitment and/or premature or sustained activation will result in destruction of healthy tissue and disease. It is now well established that most, if not all, chemokines exist as monomers and dimers, but the molecular mechanisms by which chemokines and monomer-dimer equilibrium mediate leukocyte function remain unknown. Using mouse models, cellular assays, and engineered monomers and dimers, we have shown that both monomers and dimers have differential activities and recruit neutrophils in a highly differential manner, that monomer-dimer equilibrium regulates recruitment, that monomers and dimers differentially bind in vivo GAGs, and that the role of GAG interactions is highly tissue-specific.
Structural Basis of GAG-chemokine interactions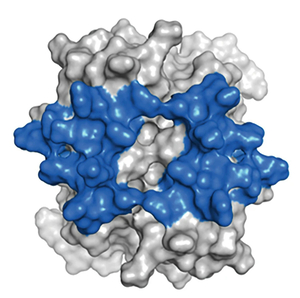 GAG-bound chemokine gradients in the blood and extracellular matrix play crucial roles in directing leukocytes to the
site of tissue infection. GAGs, such as heparan sulfate and heparin, are linear polysaccharides, and highly acidic due to multiple sulfate/carboxylate groups in the repeating disaccharide unit. Structure-function studies have established that salt
bridge interactions mediate binding. However, very little is known regarding the molecular basis by which these interactions impart specificity, affinity, and function. Our lab is actively studying how chemokines recognize and bind GAGs using a variety
of biophysical methods. In particular, we are interested in developing NMR probes to better describe how proteins engage GAGs, and to understand the causal relationships between structure, dynamics, thermodynamics, and function.
GAG-bound chemokine gradients in the blood and extracellular matrix play crucial roles in directing leukocytes to the
site of tissue infection. GAGs, such as heparan sulfate and heparin, are linear polysaccharides, and highly acidic due to multiple sulfate/carboxylate groups in the repeating disaccharide unit. Structure-function studies have established that salt
bridge interactions mediate binding. However, very little is known regarding the molecular basis by which these interactions impart specificity, affinity, and function. Our lab is actively studying how chemokines recognize and bind GAGs using a variety
of biophysical methods. In particular, we are interested in developing NMR probes to better describe how proteins engage GAGs, and to understand the causal relationships between structure, dynamics, thermodynamics, and function.
Structural basis of receptor activation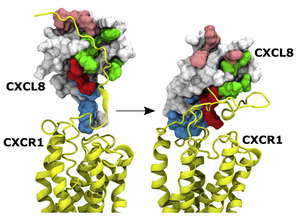 Chemokines are unconventional agonists for class A GPCRs, and the large size of chemokines
must confer functional advantages that cannot be achieved by small molecule agonists, and so must have evolved distinct molecular mechanisms that exploit their large size and multi-modular structure. All chemokines share thesame structure, but at
the functional level, no two chemokines are alike. Some chemokines are highly specific and others are promiscuous; further, monomers and dimers can bind with different selectivity and elicit a range of downstream signaling events. Receptor activation
involves interactions between chemokine N-loop and receptor N-terminal residues (defined as Site-I), and between chemokine N-terminal and receptor extracellular/transmembrane residues (defined as Site-II). How this two-site model enables large variation
in receptor selectivity, affinity, and activity is not known.We propose structural and dynamic features of chemokine N-loop and receptor N-domain residues allow a spectrum of Site-I interactions, and that Site-I and Site-II interactions are coupled
which result in differential activation of various signaling pathways in a chemokine-dependent, monomer/dimer dependent, and receptor-dependent manner. We are currently studying how neutrophil-activating chemokines bind and activate CXCR1 and CXCR2
receptors.
Chemokines are unconventional agonists for class A GPCRs, and the large size of chemokines
must confer functional advantages that cannot be achieved by small molecule agonists, and so must have evolved distinct molecular mechanisms that exploit their large size and multi-modular structure. All chemokines share thesame structure, but at
the functional level, no two chemokines are alike. Some chemokines are highly specific and others are promiscuous; further, monomers and dimers can bind with different selectivity and elicit a range of downstream signaling events. Receptor activation
involves interactions between chemokine N-loop and receptor N-terminal residues (defined as Site-I), and between chemokine N-terminal and receptor extracellular/transmembrane residues (defined as Site-II). How this two-site model enables large variation
in receptor selectivity, affinity, and activity is not known.We propose structural and dynamic features of chemokine N-loop and receptor N-domain residues allow a spectrum of Site-I interactions, and that Site-I and Site-II interactions are coupled
which result in differential activation of various signaling pathways in a chemokine-dependent, monomer/dimer dependent, and receptor-dependent manner. We are currently studying how neutrophil-activating chemokines bind and activate CXCR1 and CXCR2
receptors.