SoftwareTo obtain a copy of the software, please contact Dr. Gillian Lynch (gclynch@utmb.edu.)
ESP - Extended System Program for Molecular Dynamics
Parallel molecular dynamics code designed for simulation of large biomolecular systems. The program includes the glide-plane boundary condition for simulations of interfacial systems [K.Y. Wong and B. M. Pettitt, Chem. Phys. Lett. 326, 193 (2000)], and replica-exchange lambda sampling [C. Y. Hu, H. Kokubo, G. C. Lynch, D. W. Bolen, and B. M. Pettitt, Protein Sci. 19, 1011 (2010)]
3DRISM
Integral equations (IEs) of liquid state theory are used to determine solvent effects of molecular systems. This program includes the application of 3D-IE methods to multisite atomic models and incorporates interaction site model (ISM) theory. The electrostatic effects are included via the more exact long-range treatment of the correlation functions that uses a resummation technique [J. S. Perkyns, G. C. Lynch, J. J. Howard, and B. M. Pettitt, "Protein solvation from theory and simulation: Exact treatment of Coulomb interactions in three-dimensional theories", Journal of Chemical Physics, 132(6), 064106 (2010)]. The equations can be solved using Picard, MDIIS or Newton-type methods which were developed at the University of Houston [J. J. Howard, J. S. Perkyns, N. Choudhury, and B. M. Pettitt, "An Integral Equation Study of the Hydrophobic Interaction between Graphene Plates," Journal of Chemical Theory and Computation, 4(11), 1928 (2008)].
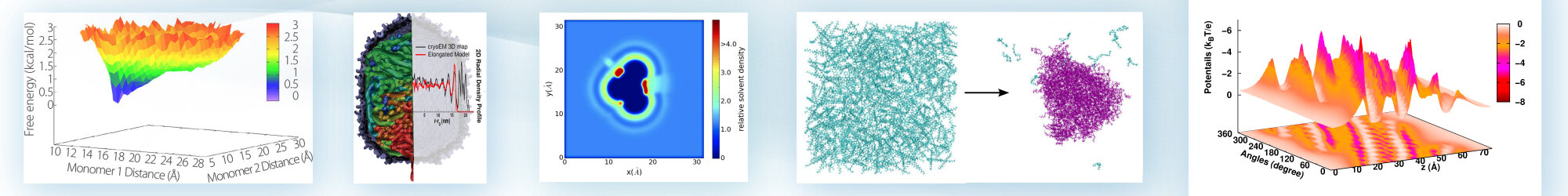
Prdf - Proximal radial distribution functions
A method, called the proximal radial distribution function (pRDF), for reconstructing the solvent density using the proximity criteria. The pRDF is hypothesized to be a universal descriptor based on a dominant local interaction between the solvent and the chemical nature of the protein atoms. The resultant distribution function can be defined as perpendicular to the surface of the molecule of interest and used for experimental and theoretical analysis.
["Note: On the universality of proximal radial distribution functions of proteins," R. Lin and B. M. Pettitt, Journal of Chemical Physics, 134, 106101 (2011)]
MST-CG - Microarray Structure and Thermodynamics - Coarse Grained Approach
Parameterization of a discretized worm-like chain model is used to determine the properties of single (ssDNA) and double stranded DNA (dsDNA) tethered to a surface. Each sphere represents 6 base pairs of a dsDNA or 1.5 base pairs for ssDNA. Configurational sampling is accomplished via Metropolis Monte-Carlo simulations. Entropy contributions are also included using two different techniques: (1) a quasi-harmonic approximation and (2) hypothetical scan method.
["A model for structure and thermodynamics of ssDNA and dsDNA near a surface: A coarse grained approach," J. Ambia-Garrido, A. Vainrub, and B. M. Pettitt, Computer Physics Communications, 181, 2001 (2010) and "Free energy considerations for nucleic acids with dangling ends near a surface: a coarse grained approach," J. Ambia-Garrido, A. Vainrub, and B. M. Pettitt, Journal of Physics: Condensed Matter, 23, 325101 (2011).]