Videos
DNA Packaging | |||||||||||||||||||||
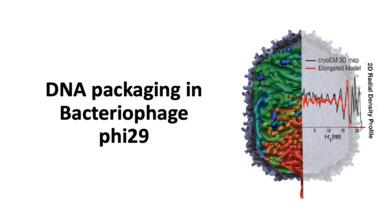 | DNA packaging in bacteriophage phi29 This video presents the work of effects of model shape, volume, and softness of the capsid for DNA packaging of phi29. Pairs of nucleotides are displaced into the capsid at a constant rate and under a torsional force. Bores C, Woodson M, Morais MC, Pettitt BM. J Phys Chem B. 124(46), 10337-10344 (2020) | ||||||||||||||||||||
DNA Pairing | |||||||||||||||||||||
 | Interactions between identical DNA double helices This video presents the work of molecular dynamics simulations on DNA-DNA interactions in 150 mM NaCl solution and to explain how the free-energy landscape of DNA-DNA interactions depends on the separation and orientation of the DNA molecule. Lai CL, Chen C, Ou SC, Prentiss M, Pettitt BM. Physical Review E 101, 032414 (2020). | ||||||||||||||||||||
Aggregation of Pentaglycines | |||||||||||||||||||||
 | Solubility and Aggregation of Gly5 in Water This video presents the large scale simulations with over 600 pentaglycines at 0.3 M concentrations in explicit solvent to consider the mechanism of aggregation. Karandur D, Wong KY, Pettitt BM. J Phys Chem B, 118(32), 9565-72 (2014). | ||||||||||||||||||||
Protein-DNA binding | |||||||||||||||||||||
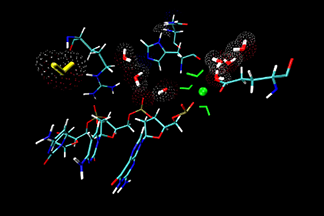 | Water migration at the active site
This video shows the solvent participation in Serratia marcescens endonuclease (SMnase)-DNA complexes. A water molecule colored yellow enters the active site of SMnase, and interacts with other water molecules, then leaves. C. Chen, et al. Proteins: Struct. Funct. and Bioinfo., 62, 982-995 (2006). | ||||||||||||||||||||
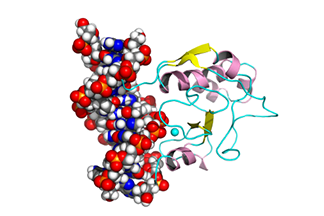 | Binding process between protein and DNA
This is an award-winning video in the VisVid Fest 2009 at the Texas Learning and Computation Center, University of Houston, Texas. It shows the binding process between the N-ColE7 protein and its DNA target. C. Chen and B.M. Pettitt. Biophys. J, 101, 1139-1147 (2011) | ||||||||||||||||||||
 | Dipole moment vector of water molecules around a DNA
DNA is highly charged biomolecule. It can interact strongly with the water molecules around it and restrict both their transitional and rotational degree of freedom. This video shows the water dipole field at the hydration sites around the DNA. The magnitudes of the dipole moments decrease in the color scale from red, yellow, green and cyan. (by C. Chen) | ||||||||||||||||||||
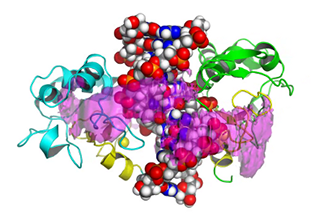 | 3D Map of binding energy between protein and DNA
This video shows three different energy contour levels (pink, orange and magenta in decreasing order) of N-ColE7 binding with the DNA duplex. The energy isosurface (magenta) indicates the formation of an encounter state could happen in either the minor groove or the major groove. And transient encounter states play an important role in a variety of dynamic process leading to molecular recognition. C. Chen and B.M. Pettitt. Biophys. J, 101, 1139-1147 (2011). | ||||||||||||||||||||
Underwinding and overwinding of DNA helices | |||||||||||||||||||||
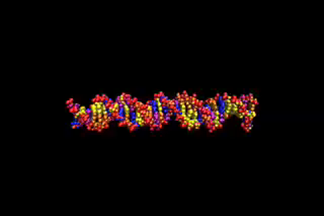 | This video is S10, one of a series from 19 molecular dynamics simulations performed to show the behavior of DNA duplex as a function of underwinding and overwinding. The bases are colored: adenine, blue; thymine, purple; guanine, yellow; and cytosine, orange. The movies are labeled S1, DNA underwound, to S19, DNA overwound, S10 is the control. Undertwisting results in strand separation in sequence specific places. P-DNA is seen for the overwound system, S19. Graham L. Randall, et al. Nucl. Acids Res., 37, 5568-5577 (2009).
| ||||||||||||||||||||
| |||||||||||||||||||||
DNA microarray | |||||||||||||||||||||
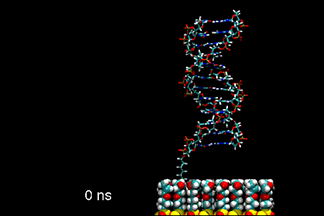 | DNA duplex tethered to a surface
This video shows a molecular dynamics simulation of a 12-base-pair DNA duplex tethered to a neutral, epoxide-coated silica surface. Starting with a canonical B-form in an up-right position, normal to the surface, the DNA tilted to over 55° and back. The linker between the DNA and the surface went from standing upright to tilted, and finally collapsed on the surface. The conformation of DNA remained closed to B-form. K.-Y. Wong, et al. Biopolymers, 73, 570-578 (2004). | ||||||||||||||||||||
Single-stranded DNA on positive-charged surface
| |||||||||||||||||||||
Melting of DNA duplex | |||||||||||||||||||||
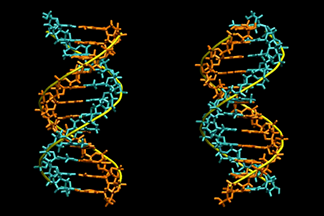 | This video shows a molecular dynamics simulation of a homo-oligomeric, 12-basepair DNA duplex, d(A12)·d(T12), with explicit salt water at 400 K. The DNA is showed in two perpendicular views and is spinned at a constant speed for illustration. It reveals the various biochemically important processes that occur on different timescales. K.-Y. Wong, et al. Biophysical Journal, 95, 5618-5626 (2008). | ||||||||||||||||||||
Parallel implementation of Fast Multipole Algorithm | |||||||||||||||||||||
 | This video was presented by J. Kurzak at Supercomputing 2004 as part of a collaboration with the Pittsburgh Supercomputer Center. It illustrates the difficult challenges and innovative solutions of implementing Fast Multiple Algorithm in molecular dynamics simulation program running on massively parallel computers. | ||||||||||||||||||||