Principal Investigator

Guy NirAssistant Professor
Department of Biochemistry & Molecular Biology
Department of Neuroscience, Cell Biology and Anatomy
Sealy Center for Structural Biology & Molecular Biophysics
In January 2021 I have joined the Department of Biochemistry and Molecular Biology, became affiliated with the Department of Neuroscience, Cell Biology and anatomy, and a member of the sealy Center for Structural Biology and Molecular Biophysics at the University of Texas Medical Branch (UTMB) in Galveston, TX.
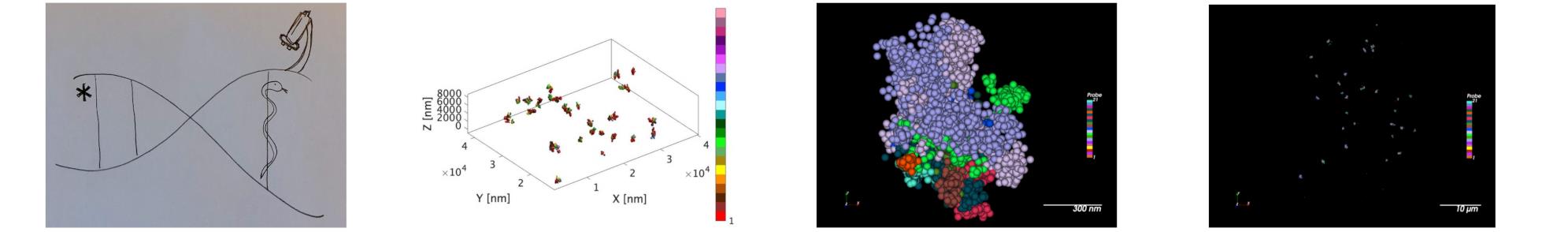
Prior to joining UTMB, I was a postdoctoral fellow in the lab of Prof. Ting Wu. During my time with Ting, I worked to describe the mechanisms of genome folding with mainly single-cell imaging technologies. At Ting’s lab, I developed a new approach to image multiple genomic targets with super-resolution microscopy. In our PLoS Genetics 2018 paper, we describe how we developed this technology, and how we used to it image several genomic targets, including a 3 kb regulatory elements and its corresponding gene (DNMT1), a loop, four contact domains, and nine chromosomal segments. Together with the lab of Marc A. Marti-Renom, and his postdoctoral fellow Irene Farabella, we developed new analysis tools, and with those, we showed that the nine chromosomal segments contributed to five spatially-segregated compartments. Through a collaboration with the lab of Erez Lieberman Aiden, and his postdoctoral fellow, Cynthia Perez-Estrada, we compared our results with new Hi-C data and showed how the imaging and Hi-C analyses were mostly in agreement. With our imaging, we also described the cell-to-cell variability, which can help in understanding the strength of each compartment. Together with both the Marti-Renom and Aiden labs, we integrated the imaging with Hi-C data and modeled the structure of the chromosomes at high genomic and spatial resolution. Finally, we detected the paternal and maternal homologs in situ and learned that these are more different than random. We also enjoyed collaborating with the lab of Peng Yin and Bruker Inc.
I have conducted my Ph.D. research in Prof. Yuval Garini lab at the physics department and Nanotechnology Institute, Bar Ilan University (BIU), Ramat-Gan, Israel. During that time I have used, developed and implemented single molecule in vitro techniques to study DNA, protein-DNA interactions and proteins unfolding dynamics.
I have described how the HU protein affects single dsDNA molecules in vitro using the Tethered Particle Motion (TPM) method. TPM is a force-free single-molecule technique that allows observing spatiotemporal conformational variations of single biopolymers, e.g. DNA and proteins. TPM differs from other single-molecule techniques, as it doesn’t require to apply force and hence, it is possible to observe single molecules in their natural form. I have then optimized TPM to provide a broader tool to probe single molecules. We are now preparing two manuscripts to describe our findings.