Structures
X-ray crystallography
Cryo-electron Microscopy and Tomography
Structures on hold for publication at PDB or EMDB
Protein Data Bank (PDB)
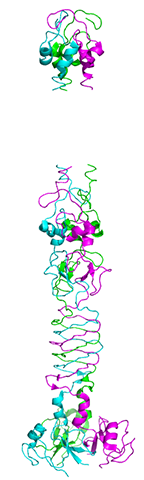
Re-refined structure of the bacteriophage T4 short tail fibre PDB entry 1H6W containing 71 additionally identified residues
Taylor NMI, van Raaij MJ, Leiman PG.
Contractile injection systems of bacteriophages and related systems. Mol. Microbiol. (2018) [PubMed]
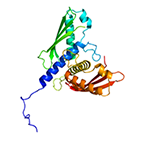
Structure of bacteriophage T4 gp25, sheath polymerization initiator
Taylor NM, Prokhorov NS, Guerrero-Ferreira RC, Shneider MM, Browning C, Goldie KN, Stahlberg H, Leiman PG.
Structure of the T4 baseplate and its function in triggering sheath contraction. Nature 533, 346-52 (2016) [PubMed]
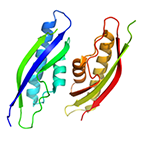
C-terminal domain of the R-type pyocin baseplate protein PA0618
Leiman PG, Plattner M, Buth SA, Shneider MM
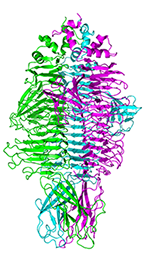
Gp54 tailspike of Acinetobacter baumannii bacteriophage AP22 in complex with A. baumannii capsular saccharide
Buth SA, Shneider MM, Leiman PG
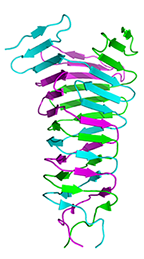
Crystal structure of the C-terminal domain of R2 pyocin membrane-piercing spike
Browning CB, Leiman PG, Shneider MM
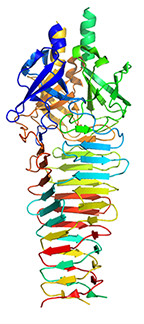
Crystal structure of R2 pyocin membrane-piercing spike
Browning CB, Leiman PG, Shneider MM
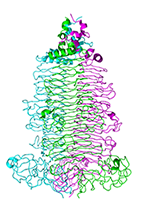
Apo crystal structure of the colanidase tailspike protein gp150 of Phage Phi92
Browning C, Schwarzer D, Gerardy-Schahn R, Shneider MM, Leiman PG
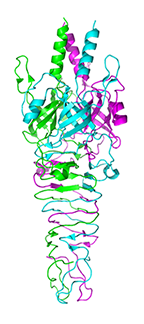
Crystal structure of the cell puncturing protein gp41 from Pseudomonas phage SN
Browning C, Shneider MM, Leiman PG
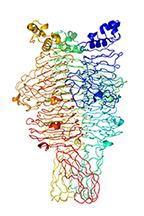
Crystal structure of the tailspike protein gp49 from Pseudomonas phage LKA1
Olszak T, Shneider MM, Latka A, Maciejewska B, Browning C, Sycheva LV, Cornelissen A, Danis-Wlodarczyk K, Senchenkova SN, Shashkov AS, Gula G, Arabski M, Wasik S, Miroshnikov KA, Lavigne R, Leiman PG, Knirel YA, Drulis-Kawa Z.
The O-specific polysaccharide lyase from the phage LKA1 tailspike reduces Pseudomonas virulence. Sci Rep 7(1):16302 (2017). [PubMed]
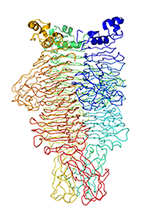
Crystal Structure of the Pseudomonas phage phi297 tailspike gp61
Olszak T, Shneider MM, Latka A, Maciejewska B, Browning C, Sycheva LV, Cornelissen A, Danis-Wlodarczyk K, Senchenkova SN, Shashkov AS, Gula G, Arabski M, Wasik S, Miroshnikov KA, Lavigne R, Leiman PG, Knirel YA, Drulis-Kawa Z.
The O-specific polysaccharide lyase from the phage LKA1 tailspike reduces Pseudomonas virulence. Sci Rep 7(1):16302 (2017). [PubMed]

Dimer of a C-terminal fragment of phage T4 gp5 beta-helix
Buth SA, Leiman PG, Shneider MM
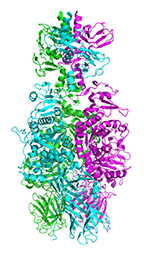
Crystal structure of tail fiber protein gp63.1 from E. coli phage G7C
Prokhorov NS, Riccio C, Zdorovenko EL, Shneider MM, Browning C, Knirel YA, Leiman PG, Letarov AV
Function of bacteriophage G7C esterase tailspike in host cell adsorption. Mol. Microbiol. 105, 385-398 (2017) [PubMed]
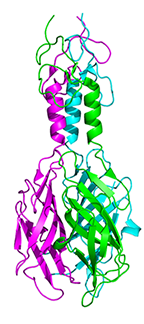
Crystal structure of the tail fiber gp53 from Acinetobacter baumannii bacteriophage AP22
Sycheva LV, Shneider MM, Popova AV, Ziganshin RK, Miroshnikov KA, Leiman, PG
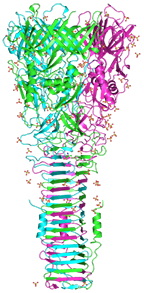
Crystal structure of PA0091 VgrG1, the central spike of the Type VI Secretion System
Sycheva LV, Shneider M, Basler M, Ho BT, Mekalanos JJ, Leiman PG

Phage T4 spike tip protein gp5.4 in complex with T4 gp5 beta-helix
Buth SA, Shneider MM, Leiman PG
Gp5.4 amino acid sequence and gene information is here. Gp5 amino acid sequence and gene information is here (residues 484-575).
Gp5 and gp5.4 were co-expressed in cis. Expression construct type: His-tag_TEV-cleavage-site_gp5...gp5.4.
Expression vector: pEEva2 (pET-23a derivate).
Structure and biophysical properties of phage T4 gp5 beta-helix
Buth SA, Leiman PG, Boudko SP
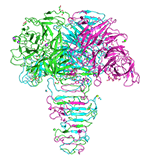
Catalytic domain of phage phi92 endosialidase (residues 76-756 of gp143)
Schwarzer D, Browning C, Leiman PG
Amino acid sequence and gene information is here
Expression construct type: Strep-tag_Thrombin-cleavage-site_gp143(aa76-918)_His-tag
Expression vector: pET22b-Strep (pET22b with N-terminal Strep-tag)

PAAR-repeat binding tip (residues 561-633) of the Z0707 VgrG protein of E. coli EDL933
Uchida K, Leiman PG, Arisaka F, Kanamaru S
Structure and properties of the C-terminal β-helical domain of VgrG protein from Escherichia coli O157. J. Biochem. 155, 173-182 (2013). [PubMed]
Amino acid sequence and gene information is here
Expression construct type: His-tag_SlyD-TEV-cleavage-site_z0707
Expression vector: pESL (pET28a derivative)
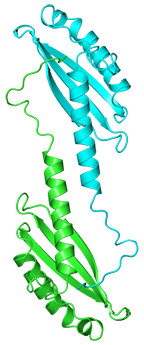
Phage T4 baseplate protein gp25
Browning C, Shneider MM, Leiman PG
Amino acid sequence is here
Expression construct type: His-tag_SlyD-TEV-cleavage-site_gp25
Expression vector: pESL (pET28a derivative)

c1882 PAAR-repeat protein of E. coli CFT073 in complex with T4 gp5-VgrG chimera
Shneider MM, Buth SA, Ho BT, Basler M, Mekalanos JJ, Leiman PG
PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature. 500, 350-353 (2013). [PubMed]
Gp5 amino acid sequence is here (residues 484-575). Expression construct type: His-tag_TEV-cleavage-site_gp5. Expression vector: pEEva2 (pET-23a derivate).
c1882 amino acid sequence is here. Expression construct type: untagged. Expression vector: pATE (pACYCDuet-1 derivative).
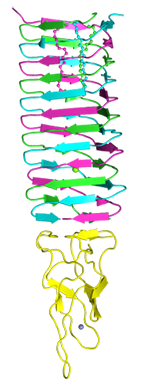
VCA0105 PAAR-repeat protein of V. cholerae O1 biovar El Tor str. N16961 in complex with T4 gp5-VgrG chimera
Shneider MM, Buth SA, Ho BT, Basler M, Mekalanos JJ, Leiman PG
PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature. 500, 350-353 (2013). [PubMed]
Gp5 amino acid sequence is here (residues 484-575). Expression construct type: His-tag_TEV-cleavage-site_gp5. Expression vector: pEEva2 (pET-23a derivate).
VCA0105 amino acid sequence is here. Expression construct type: untagged. Expression vector: pATE (pACYCDuet-1 derivative).
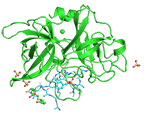
Human urokinase-type plasminogen activator uPA with bound bicyclic peptide inhibitor UK18-D-Aba
Chen S, Gfeller D, Buth SA, Michielin O, Leiman PG, Heinis C
Improving Binding Affinity and Stability of Peptide Ligands by Substituting Glycines with D-Amino Acids. Chembiochem. 14, 1316-1322 (2013). [PubMed]
Amino acid sequence and gene information is here.
Commercially produced protein purchased here.
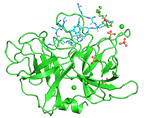
Human urokinase-type plasminogen activator uPA with bound bicyclic peptide inhibitor UK18-D-Ser
Chen S, Gfeller D, Buth SA, Michielin O, Leiman PG, Heinis C
Improving Binding Affinity and Stability of Peptide Ligands by Substituting Glycines with D-Amino Acids. Chembiochem. 14, 1316-1322 (2013). [PubMed]
Amino acid sequence and gene information is here.
Commercially produced protein purchase here.
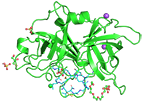
Human urokinase-type plasminogen activator uPA with bound bicyclic peptide inhibitor UK504
Chen S, Rentero Rebollo I, Buth SA, Morales-Sanfrutos J, Touati J, Leiman PG, Heinis C
Bicyclic Peptide Ligands Pulled out of Cysteine-Rich Peptide Libraries. J. Am. Chem. Soc. 135, 6562-6569. (2013). [PubMed]
Amino acid sequence and gene information is here.
Commercially produced protein purchased here.
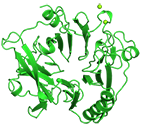
C-terminal fragment of phage phiKZ tail fiber protein gp131 (residues 375-771)
Sycheva LV, Shneider MM, Sykilinda NN, Ivanova MA, Miroshnikov KA, Leiman PG
Crystal structure and location of gp131 in the bacteriophage phiKZ virion. Virology. 434, 257-264 (2012). [PubMed]
Amino acid sequence and gene information is here.
Expression construct type: His-tag_SlyD-TEV-cleavage-site_gp131
Expression vector: pESL (pET28a derivative)
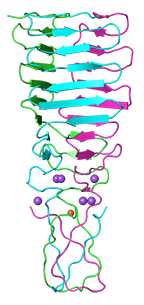
C-terminal fragment of phage phi92 central spike protein gp138 (residues 121-245)
Browning C, Shneider MM, Bowman VD, Schwarzer D, Leiman PG
Phage pierces the host cell membrane with the iron-loaded spike. Structure. 20:326-339 (2012). [PubMed]
Amino acid sequence and gene information is here.
Expression construct type: His-tag_TEV-cleavage-site_gp138C
Expression vector: pEEVa2 (pET-23a derivate
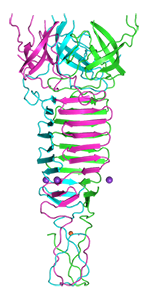
Phage phi92 central spike protein gp138
Browning C, Shneider MM, Bowman VD, Schwarzer D, Leiman PG
Phage pierces the host cell membrane with the iron-loaded spike. Structure. 20:326-339 (2012). [PubMed]
Amino acid sequence and gene information is here.
Expression construct type: His-tag_TEV-cleavage-site_gp138
Expression vector: pEEVa2 (pET-23a derivate)

C-terminal fragment of phage P2 central spike protein gpV (residues 97- 211)
Browning C, Shneider MM, Bowman VD, Schwarzer D, Leiman PG
Phage pierces the host cell membrane with the iron-loaded spike. Structure. 20:326-339 (2012). [PubMed]
Amino acid sequence and gene information is here.
Expression construct type: His-tag_TEV-cleavage-site_gpVC
Expression vector: pEEVa2 (pET-23a derivate)
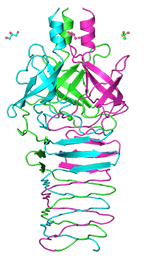
Phage P2 central spike protein gpV
Browning C, Shneider MM, Bowman VD, Schwarzer D, Leiman PG
Phage pierces the host cell membrane with the iron-loaded spike. Structure. 20:326-339 (2012). [PubMed]
Amino acid sequence and gene information is here.
Expression construct type: His-tag_TEV-cleavage-site_gpV
Expression vector: pEEVa2 (pET-23a derivate)
Electron Microscopy Data Bank (EMDB)

Cryo-EM structure of the T4 tail tube
Zheng W, Wang F, Taylor NMI, Guerrero-Ferreira RC, Leiman PG, Egelman EH
Refined Cryo-EM Structure of the T4 Tail Tube: Exploring the Lowest Dose Limit. Structure 25, 1436-1441.e2 (2017) [PubMed]
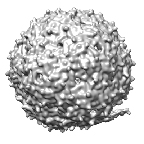
Asymmetric reconstruction of bacteriophage MS2 mutant NEO1
Zhong Q, Carratala A, Nazarov S, Guerrero-Ferreira RC, Piccinini L, Bachmann V, Leiman PG, Kohn T
Genetic, Structural, and Phenotypic Properties of MS2 Coliphage with Resistance to ClO2 Disinfection. Environ. Sci. Technol. 50, 13520-13528 (2016) [PubMed]
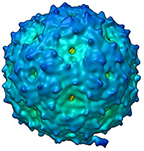
Asymmetric reconstruction of bacteriophage MS2
Zhong Q, Carratala A, Nazarov S, Guerrero-Ferreira RC, Piccinini L, Bachmann V, Leiman PG, Kohn T
Genetic, Structural, and Phenotypic Properties of MS2 Coliphage with Resistance to ClO2 Disinfection. Environ. Sci. Technol. 50, 13520-13528 (2016) [PubMed]

Cryo-electron microscopy structure of the hexagonal pre-attachment T4 baseplate-tail tube complex
Taylor NMI, Prokhorov NS, Guerrero-Ferreira RC, Shneider MM, Browning C, Goldie KN, Stahlberg H, Leiman PG
Structure of bacteriophage T4 baseplate and its function in triggering sheath contraction. NATURE 533, 346-52 (2016) [PubMed]
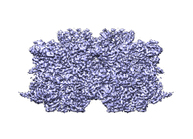
Cryo-electron microscopy structure of the hexagonal pre-attachment T4 baseplate-tail tube complex: locally masked refinement of the inner baseplate
Taylor NMI, Prokhorov NS, Guerrero-Ferreira RC, Shneider MM, Browning C, Goldie KN, Stahlberg H, Leiman PG
Structure of bacteriophage T4 baseplate and its function in triggering sheath contraction. NATURE 533, 346-52 (2016) [PubMed]
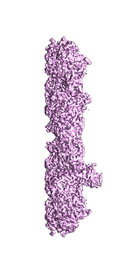
Cryo-electron microscopy structure of the hexagonal pre-attachment T4 baseplate-tail tube complex: locally masked refinement of the intermediate baseplate
Taylor NMI, Prokhorov NS, Guerrero-Ferreira RC, Shneider MM, Browning C, Goldie KN, Stahlberg H, Leiman PG
Structure of bacteriophage T4 baseplate and its function in triggering sheath contraction. NATURE 533, 346-52 (2016) [PubMed]
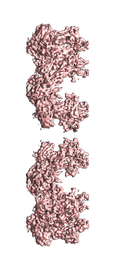
Cryo-electron microscopy structure of the hexagonal pre-attachment T4 baseplate-tail tube complex: locally masked refinement of the upper peripheral baseplate
Taylor NMI, Prokhorov NS, Guerrero-Ferreira RC, Shneider MM, Browning C, Goldie KN, Stahlberg H, Leiman PG
Structure of bacteriophage T4 baseplate and its function in triggering sheath contraction. NATURE 533, 346-52 (2016) [PubMed]
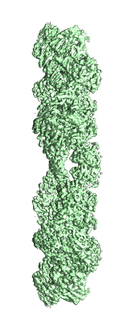
Cryo-electron microscopy structure of the hexagonal pre-attachment T4 baseplate-tail tube complex: locally masked refinement of the lower peripheral baseplate
Taylor NMI, Prokhorov NS, Guerrero-Ferreira RC, Shneider MM, Browning C, Goldie KN, Stahlberg H, Leiman PG
Structure of bacteriophage T4 baseplate and its function in triggering sheath contraction. NATURE 533, 346-52 (2016) [PubMed]

Cryo-electron microscopy structure of the star-shaped, hubless post-attachment T4 baseplate
Taylor NMI, Prokhorov NS, Guerrero-Ferreira RC, Shneider MM, Browning C, Goldie KN, Stahlberg H, Leiman PG
Structure of bacteriophage T4 baseplate and its function in triggering sheath contraction. NATURE 533, 346-52 (2016) [PubMed]

Cryo-electron microscopy structure of the hexagonal pre-attachment T4 baseplate-tail tube complex: locally masked refinement of the tail tube
Taylor NMI, Prokhorov NS, Guerrero-Ferreira RC, Shneider MM, Browning C, Goldie KN, Stahlberg H, Leiman PG
Structure of bacteriophage T4 baseplate and its function in triggering sheath contraction. NATURE 533, 346-52 (2016) [PubMed]

Atomic structures of a bactericidal contractile nanotube in its pre- and post-contraction states
Ge P, Scholl D, Leiman PG, Yu X, Miller JF, Zhou ZH
Atomic structures of a bactericidal contractile nanotube in its pre- and post-contraction states. NAT.STRUCT.MOL.BIOL. 22, 377-382 (2015) [PubMed]

Atomic structures of a bactericidal contractile nanotube in its pre- and post-contraction states
Ge P, Scholl D, Leiman PG, Yu X, Miller JF, Zhou ZH
Atomic structures of a bactericidal contractile nanotube in its pre- and post-contraction states. NAT.STRUCT.MOL.BIOL. 22, 377-382 (2015) [PubMed]

Cryo-electron microscopy structure of bacteriophage phi92 tail
Nazarov S, Bowman VD, Leiman PG
Phi92 genome is here.
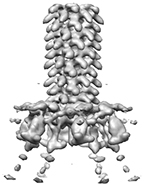
Cryo-electron microscopy structure of bacteriophage phi92 baseplate
Browning C, Nazarov S, Bowman VD, Leiman PG
Phi92 genome is here.

Cryo-electron microscopy structure of bacteriophage phi92 capsid
Browning C, Nazarov S, Bowman VD, Leiman PG
Phi92 genome is here.
PDB 4RMY
Colanic acid-bound crystal structure of the colanidase tailspike protein gp150 of Phage Phi92
Browning, C., Schwarzer, D., Gerardy-Schahn, R., Shneider, M.M., Leiman, P.G.