Research
Our research field is biophysical chemistry of proteins, DNA, and their interactions. We are working on three areas of research: 1) Biomolecular Electrostatics, 2) Protein-DNA Dynamics, and 3) HMGB1 Biochemistry.
Protein-DNA Dynamics
Proteins and DNA molecules are dynamic in many aspects. Using nuclear magnetic resonance (NMR) spectroscopy as our primary tool for research, we study various dynamics of proteins and DNA.
Dynamics of DNA scanning by proteins
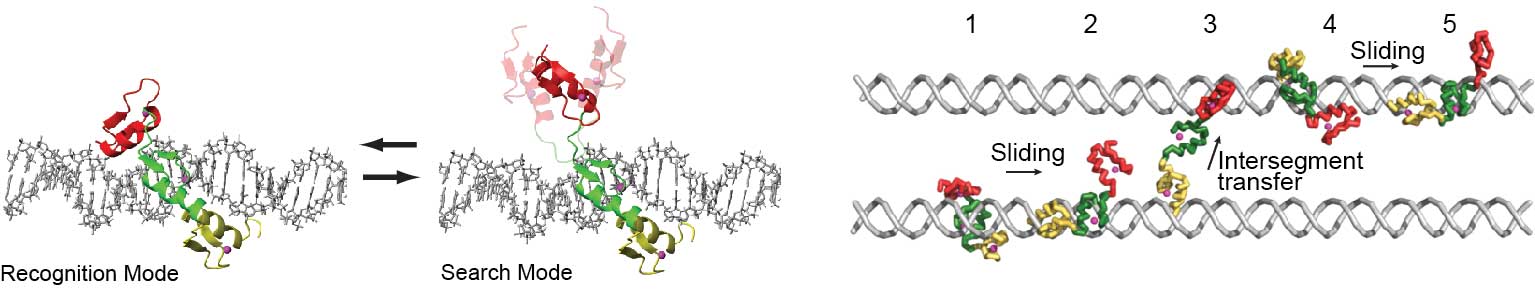
When transcription factors and DNA repair/modifying enzymes perform their function, these molecules must first locate their specific target sites in the vast presence of nonspecific but structurally similar sites on DNA. How do proteins scan DNA? To elucidate DNA-scanning mechanisms at both the molecular and atomic levels, we use various biophysical approaches. Recently we developed unique spectroscopic methods for investigations of the DNA-scanning process. We use NMR spectroscopy to characterize the DNA-scanning process at an atomic level. We also use stopped-flow fluorescence spectroscopy to study the target search kinetics at a molecular level under various conditions. Using these methodologies, we study how proteins scan DNA.
Counterion dynamics
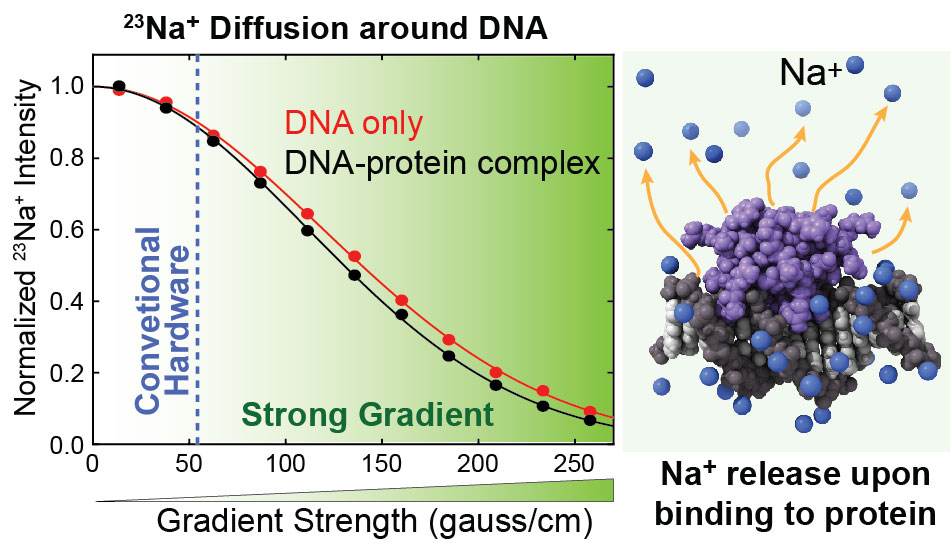
By NMR, we study how counterions behave around proteins and DNA. The large net charge on the surface of nucleic acids electrostatically attract and condense cations, creating a zone called the ion atmosphere. Experimental approaches for quantitative investigations of cations in the ion atmosphere have been developed. The counterions were found to rapidly diffuse within the ion atmosphere. Some of the counterions are released from the ion atmosphere when nucleic acids bind to proteins, neutralizing the charge via intermolecular ion pairs of positively charged side chains and negatively charged backbone phosphates. Previously, the release of counterions had only indirectly been implicated by salt-concentration dependence of the equilibrium constants for molecular association. Recently, the direct detection of the release of counterions has become possible through spectroscopic observation of ions. This allows more accurate and quantitative analysis of the counterion release and its entropic impact on the thermodynamics of protein-DNA association.
Ion-pair dynamics
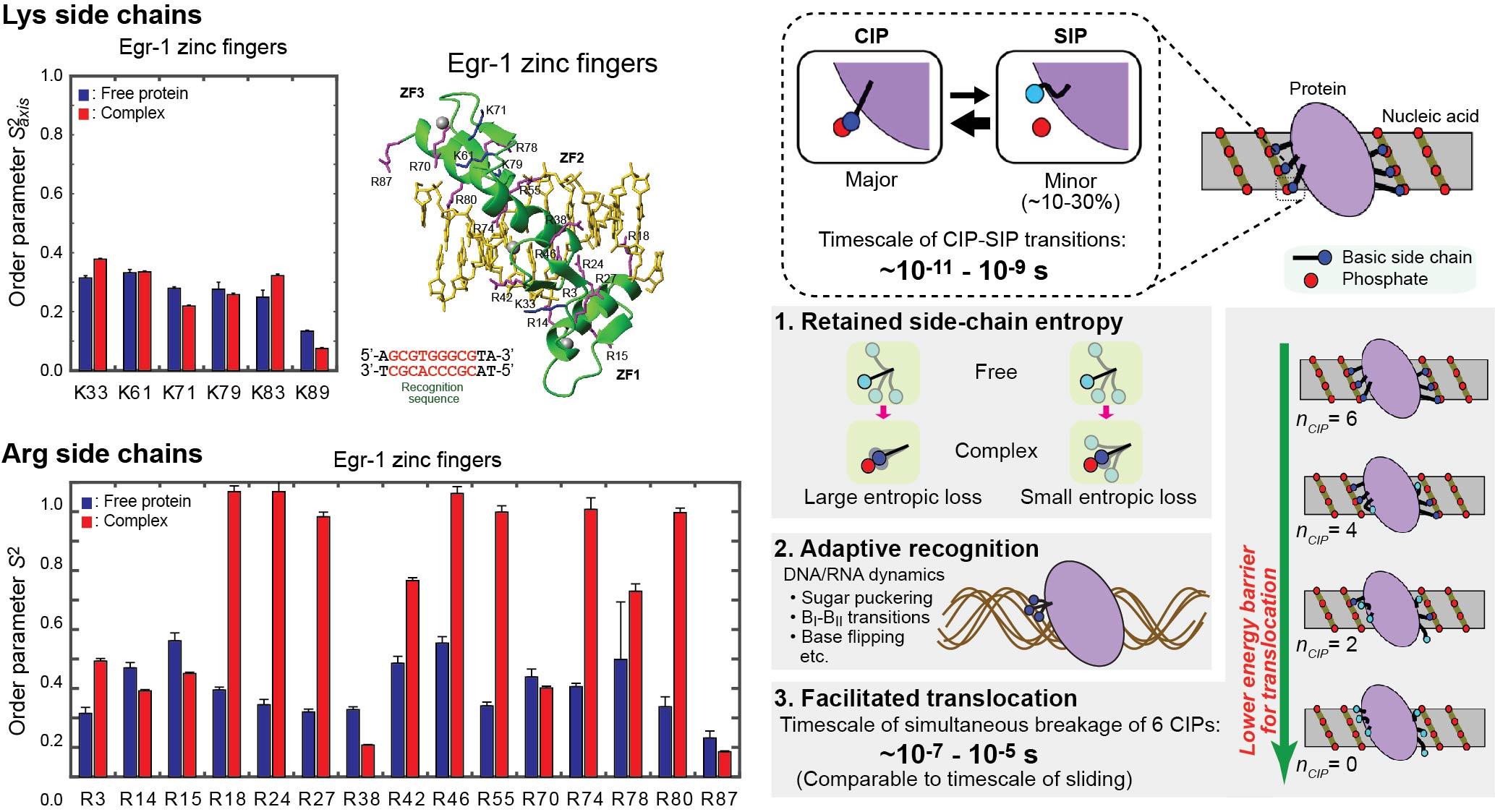
Using NMR and other biophysical methods, we also study the dynamic properties of ion pairs of protein side chains and DNA phosphates. The ion pairs undergo transitions between two major states. In one of the major states, the cation and the anion are in direct contact. This state is called a contact ion pair (CIP). In the other major state, the cation and the anion are intervened by water. This state is called a solvent-separated ion pair (SIP). Transitions between CIP and SIP states rapidly occur at the molecular interfaces. When proteins interact with nucleic acids, interfacial arginine (Arg) and lysine (Lys) side chains exhibit considerably different behaviors. Compared to Lys side chains, Arg side chains exhibit a higher propensity to directly interact with nucleotide bases, partly due to stronger cation-π interactions and a smaller desolvation energy. Lys side chains tend to be more mobile at the molecular interfaces. The dynamic ionic interactions may facilitate adaptive molecular recognition and play thermodynamic and kinetic roles in protein-nucleic acid interactions.
Selected publications