Research
Our research field is biophysical chemistry of proteins, DNA, and their interactions. We are working on three areas of research: 1) Biomolecular Electrostatics, 2) Protein-DNA Dynamics, and 3) HMGB1 Biochemistry.
HMGB1 Biochemistry
HMGB1 is a very interesting DNA-binding protein that plays important roles in the cell nuclei as well as in extracellular space. HMGB1 typically exists in the cell nuclei and serves as a DNA chaperone and assists various other DNA-binding proteins. HMGB1 contains two DNA-binding domains and intrinsically disordered regions (IDRs). HMGB1 recognizes atypical DNA such as Holliday junction, bulged DNA, cisplatin-modified DNA, and G-quadruplex. HMGB1 is released to extracellular space not only passively from dying cells, but also actively from platelets and some healthy cells. Extracellular HMGB1 serves as a danger signal and plays several important roles in innate immunity involving extracellular DNA related to infection or tissue damage. HMGB1 is involved in many inflammatory diseases and has been regarded as a potential therapeutic target. Using NMR spectroscopy, fluorescence spectroscopy, and other techniques, we are characterizing HMGB1 and trying to find effective ways for HMGB1 inhibition.
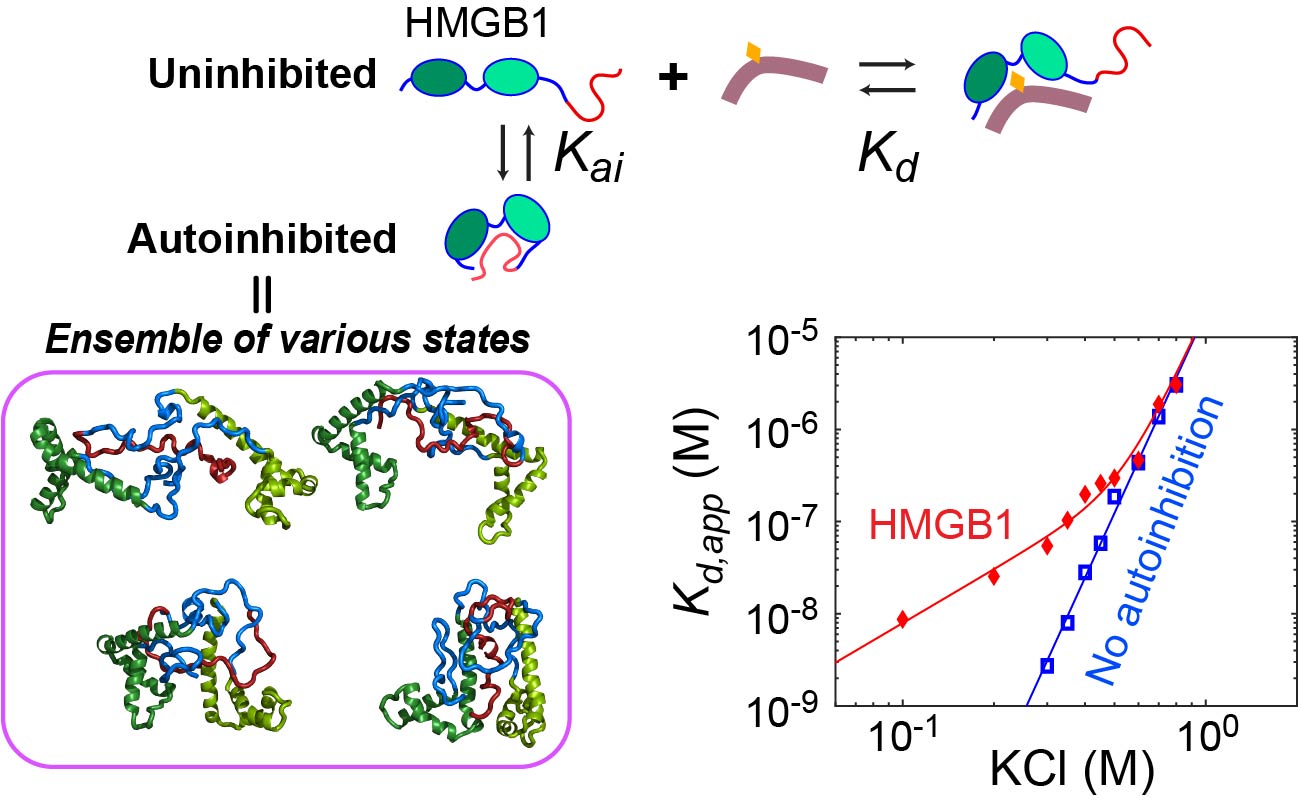
Dynamic autoinhibition
The C-terminal 30 residues of HMGB1 is a highly negatively charged IDR that contains only aspartate (D) or glutamate (E) residues. The D/E repeats interact with positively charged DNA-binding domains and cause a decrease in binding affinity for DNA. This autoinhibition occurs in a dynamic fashion involving many fuzzy interactions. Using NMR spectroscopy and fluorescence spectroscopy, we study how the autoinhibition impacts the function of HMGB1. For example, we found that the dynamic autoinhibition accelerates the target search kinetics of HMGB1 in the presence of decoys. Insight into the autoinhibition allows us to better understand the function of HMGB1.
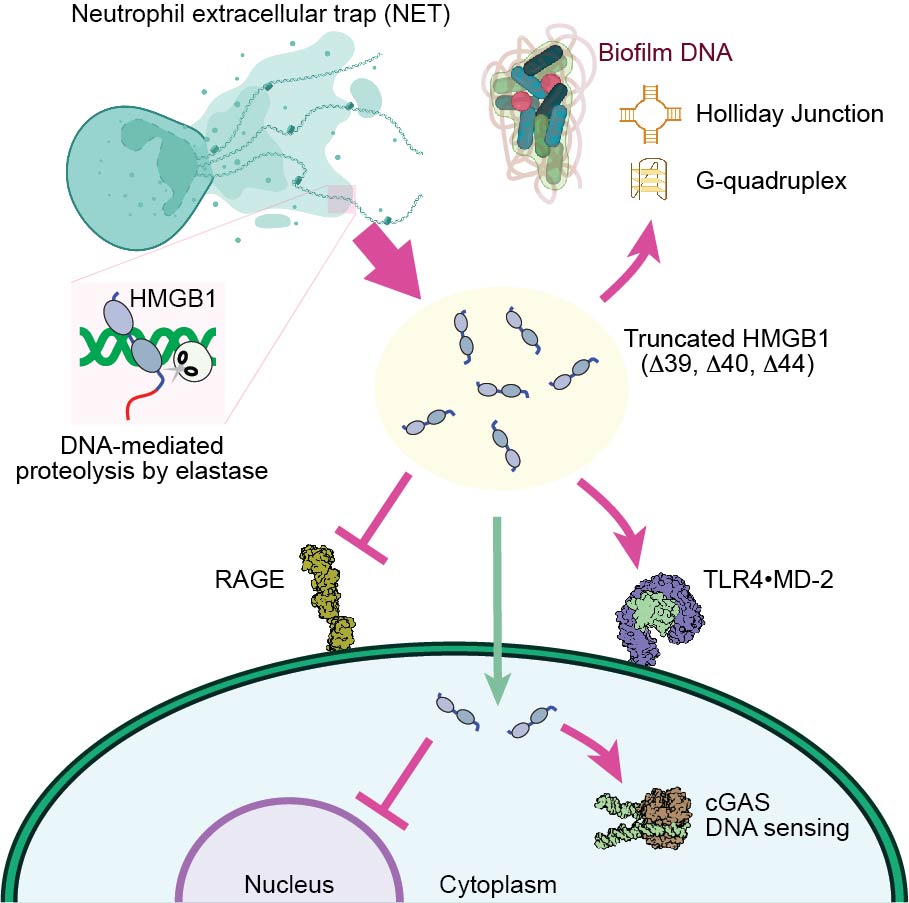
HMGB1 processing
There are several natural biochemical mechanisms that alter HMGB1's functions. Redox is one of the most important mechanisms. Extracelleular HMGB1 forms a disulfide bond between Cys23 and Cys45 and gain strong inflammatory activity. But the inflammatory activity is lost upon sulfonation of Cys106. Autoinhibition via D/E repeats is eliminated through proteolytic removal of D/E repeats via neutrophil elastase. Using various techniques, we study the impact of HMGB1 processing. Insight into the HMGB1 processing will help us develop strategies for HMGB1 inhibition.
Selected publications