Research
Our research field is biophysical chemistry of proteins, DNA, and their interactions. We are working on three areas of research: 1) Biomolecular Electrostatics, 2) Protein-DNA Dynamics, and 3) HMGB1 Biochemistry.
Biomolecular Electrostatics
In life, complex networks of molecular interactions involve electrostatic forces that influence structure and function of biological macromolecules. Electrostatics is important for our fundamental understanding of biomolecular functions as well as for drug development. Biomolecular electrostatics has been a subject of computational investigations based on 3D structures. Our nuclear magnetic resonance (NMR) methods are changing this situation. These solution NMR methods allow us to quantitatively investigate biomolecular electrostatics without any use of structure information.
Direct measurement of electrostatic potentials by NMR
We developed an innovative methodology to directly measure local electrostatic potentials for every residue of proteins and nucleic acids. The new electrostatic methods are applicable to various processes, including macromolecular association and liquid-liquid phase separation. Applications to intrinsically disordered proteins and conformationally flexible nucleic acids are particularly useful because structure-based analysis of electrostatics is not straightforward for such molecules. These tools also facilitate examination of theoretical models on biomolecular electrostatics.
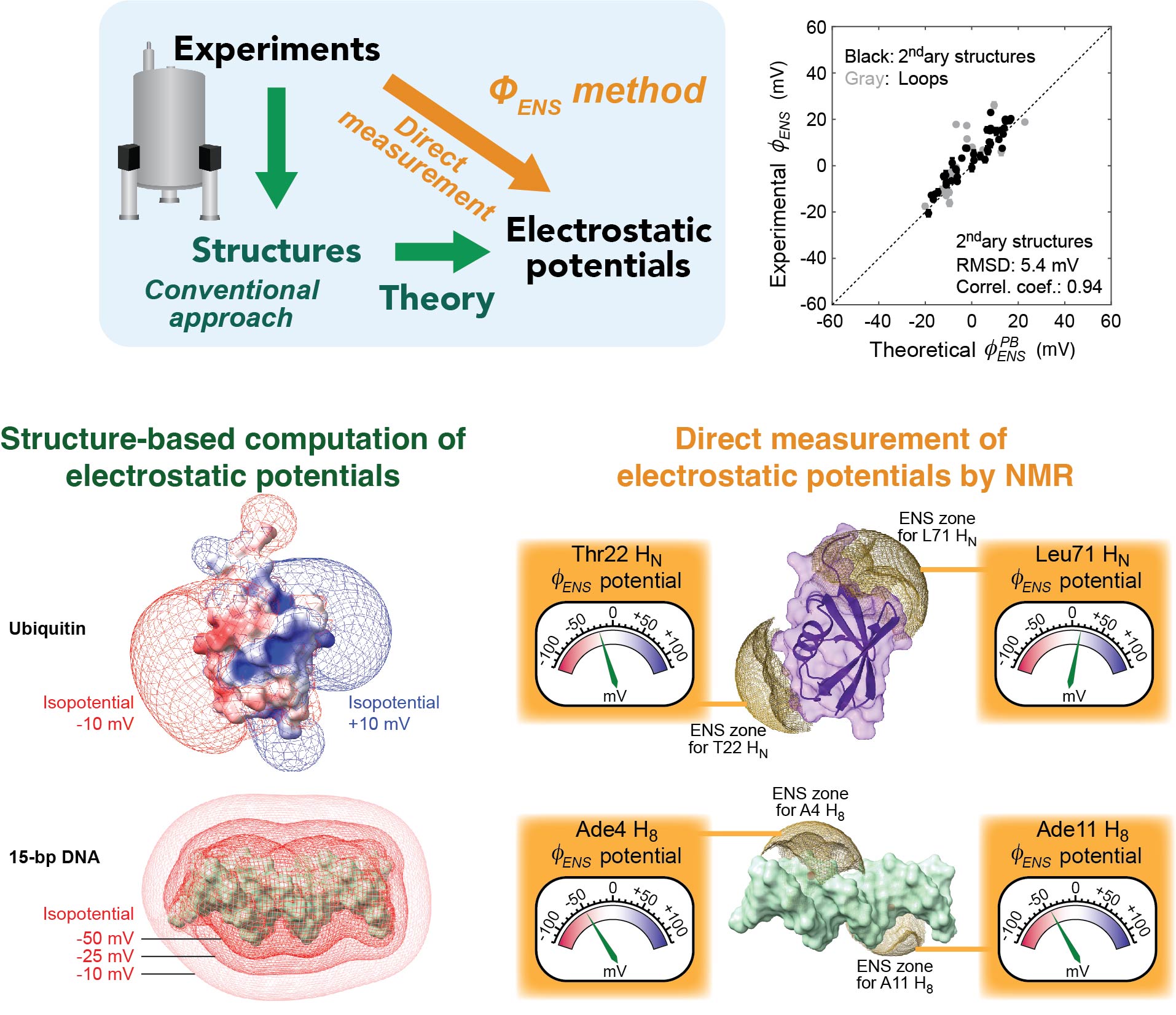
Investigations of mobile ions around biomolecules
Cellular fluids contain various inorganic and organic ions, which are essential for life. These ions electrostatically influence thermodynamics and kinetics of macromolecular interactions. Despite the importance of ions as essential constituents of biological systems, how these ions interact with biological macromolecules and impact their functions is not well understood. Mobile ions around macromolecules are difficult to analyze. Due to their mobility broadening their probability distributions, the vast majority of counterions around proteins, nucleic acids, and their complexes are unresolved even in high-resolution crystal structures. We developed NMR methods to quantitatively investigate counterions around proteins and DNA.Our NMR methods were successfully applied to 15NH4+, 23Na+, 7Li+, 133Cs+, and 13C acetate (OAc-) ions around DNA and proteins. Using the NMR methods, we study how the ions impact the functions of biomolecules.
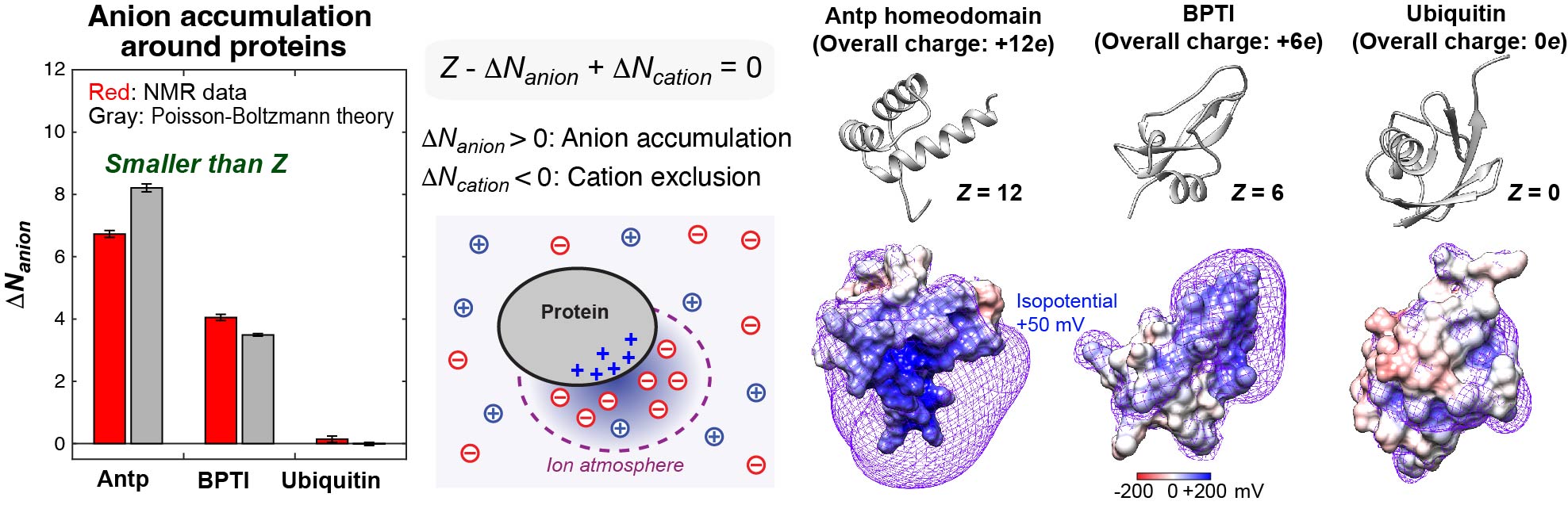
Characterizations of charged moieties of biomolecules
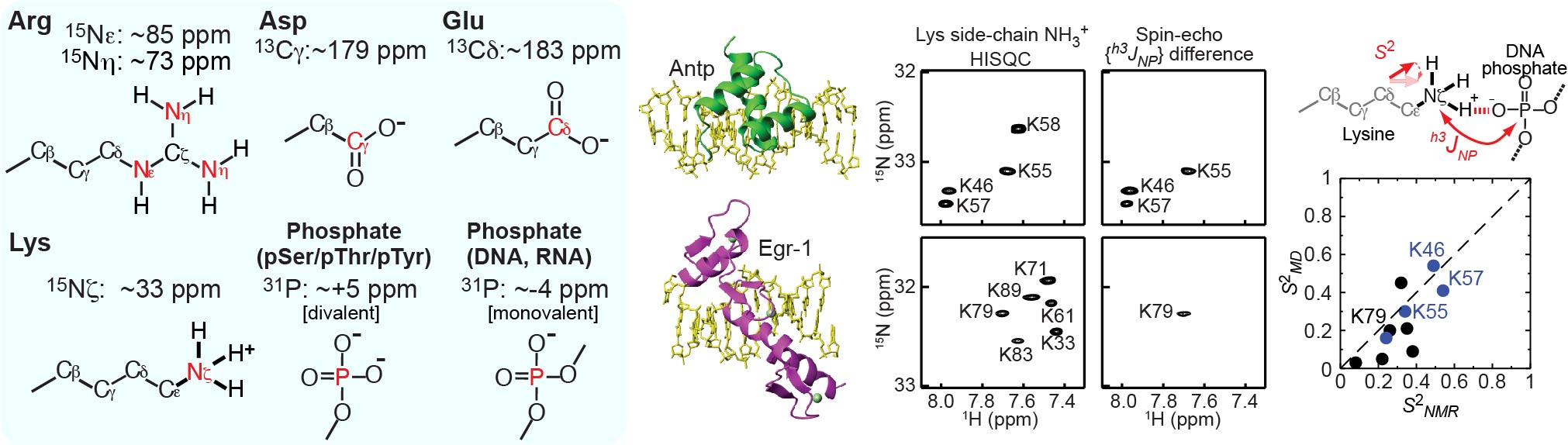
To better understand electrostatic interactions of biomolecules, characterizations of their charged moieties are essential. We use NMR spectroscopy for such characterizations of the positively or negatively charged groups of proteins and nucleic acids. 13C, 15N, and 31P nuclei that are useful for such studies: namely, 13C nuclei of aspartate/glutamate carboxylates, 15N nuclei arginine guanidinium cations, 15N nuclei of lysine amino cations; and 31P nuclei of phosphate anions of DNA, RNA, and phosphorylated protein side chains. We developed many NMR methods for Lys side-chain NH3+ groups, for example. By NMR, we learn about their electrostatic interactions, hydrogen bonds, pKa shifts, and mobilities.
Selected publications