Our Research
-
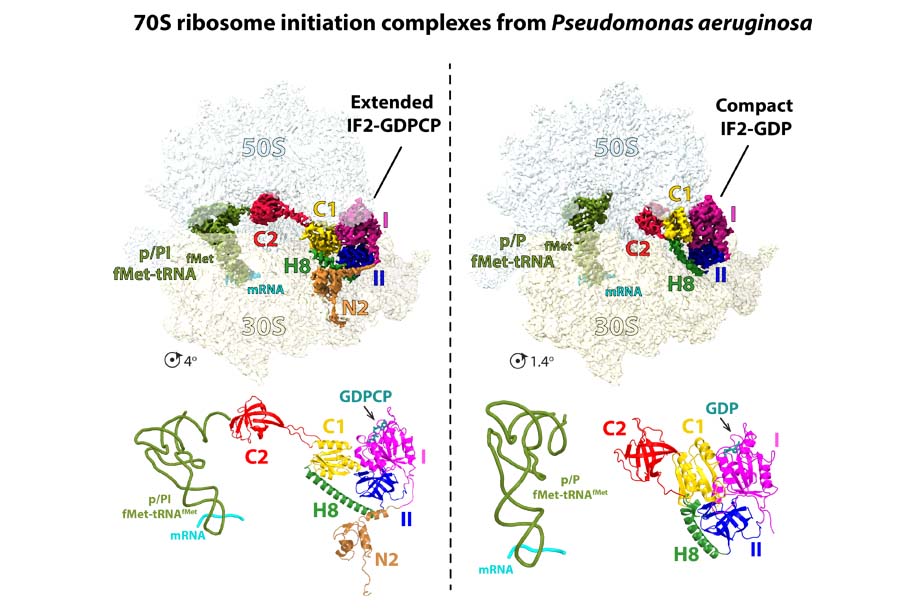
Regulation of translation initiation
In all bacteria, initiation of protein synthesis on the ribosome requires the assembly of the small 30S ribosomal subunit initiation complex at the AUG start codon. This is then followed by the association of the large 50S ribosomal subunit. Three initiation factors are known to regulate this process, IF1, IF2, and IF3. The molecular basis of the interplay between the three initiation factors, the initiator tRNA, and the ribosome, remains unclear. We recently visualized a novel compact conformation of the GTPase IF2 bound to GDP on the ribosome. By comparing with the GTP-bound form of IF2, the cryo-EM structures elucidated how IF2 changes conformation to regulate the progression of the 70S ribosome initiation complex to an elongation competent ribosome.
-
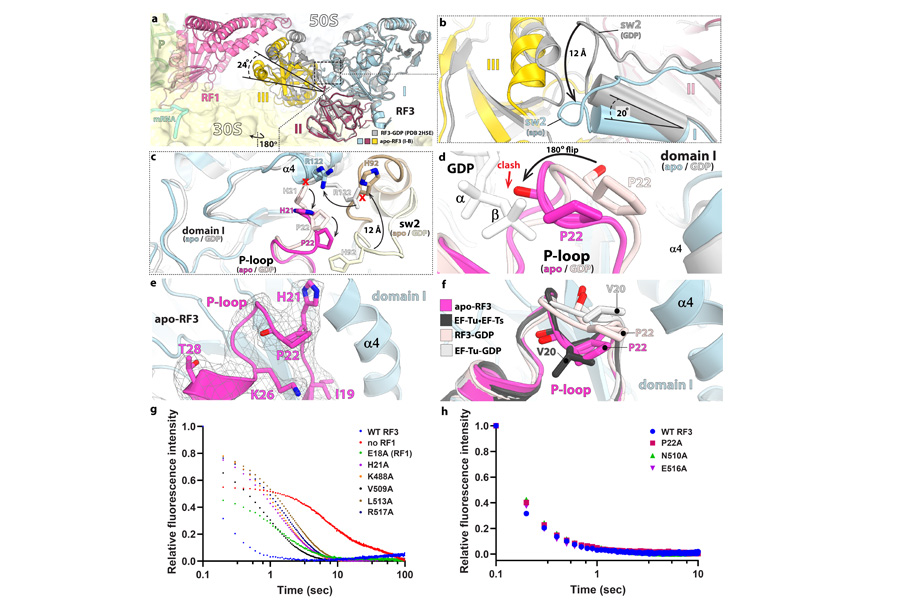
Structural bases of translation termination
In bacteria, two decoding factors recognize the three stop codons UAA, UAG, and UGA. After hydrolysis of the nascent peptide chain by RF1 or RF2, the GTPase release factor RF3 uses GTP to promote the dissociation of RF1 and RF2 from the ribosome. Studies have shown that the ribosome termination complex bound to RF1 or RF2 catalyzes the exchange of GDP for GTP in RF3. Using stopped-flow kinetics and cryo-EM, we elucidated the long-sought mechanism by which the 70S ribosome bound to RF1 promotes long-range allosteric conformational changes that remodel RF3 to eject GDP from the G-domain. We found that the guanine nucleotide exchange factor (GEF) activity of the ribosome is highly similar to that of EF-Ts, the well-characterized GEF for elongation factor-Tu (EF-Tu).
-
Molecular mechanisms of ribosome recycling
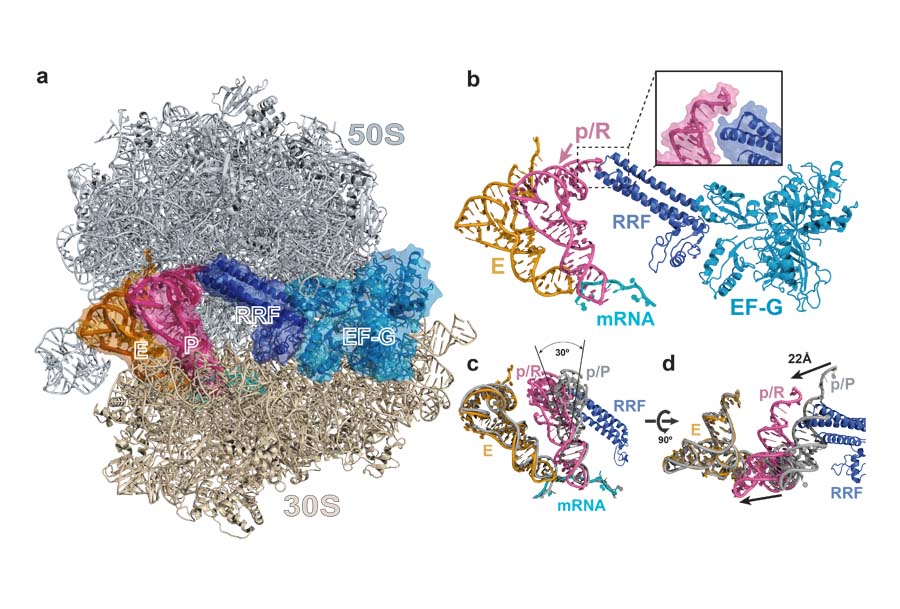
We used X-ray crystallography to determine the structure of a Thermus thermophilus 70S ribosome pre-recycling complex. Ribosome recycling is mediated by the action of the elongation factor G (EF-G), the GTPase that translocates tRNAs and mRNA by one codon every elongation cycle, and the ribosome recycling factor (RRF). We trapped EF-G in a compact conformation bound to a ribosome complexed with tRNAs and RRF. This study revealed a previously unseen state of tRNA binding in the peptidyl (P) site of the ribosome. We named it the p/R state of binding, illustrating that this conformation is induced by the simultaneous binding of RRF to the ribosome. In this conformation, the CCA-end of the p/R tRNA may help to dissociate the ribosomal subunits during ribosome recycling.
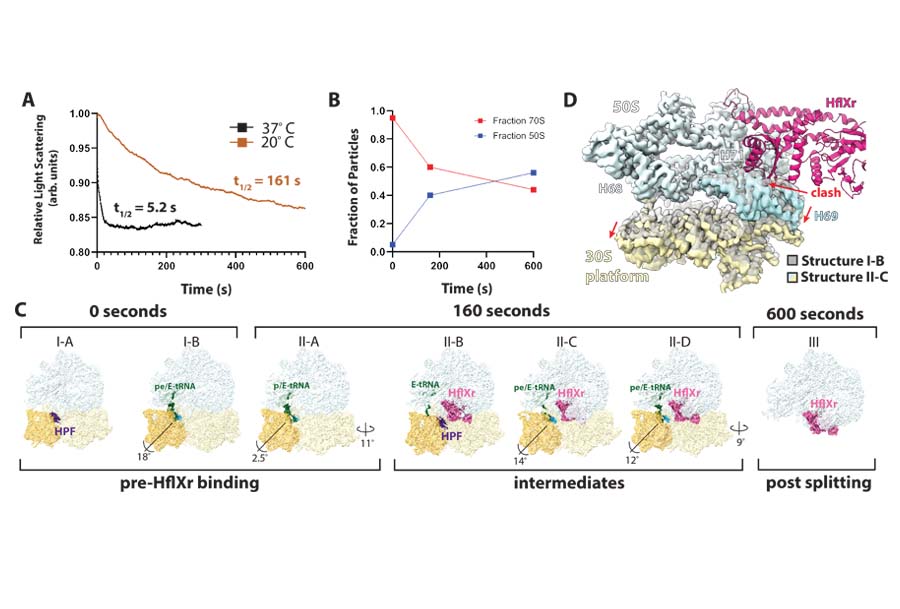
Under conditions of stress, bacteria use additional factors to recycle their stalled ribosomes. HflX is one such factor, a GTPase that promotes ribosome dissociation into individual subunits. In select bacteria, such as Listeria, an ortholog of HflX was identified and shown to be expressed under antibiotic exposure. The observation that it confers resistance to antibiotics targeting the peptidyl-transferase center (PTC) of the ribosome lead to its name, HflXr (“r” for resistance). We elucidated how HflXr promotes ribosome recycling by time-resolved cryo-EM guided by stopped-flow fast kinetic experiments.
-
Structural basis for translational control by modified nucleotides
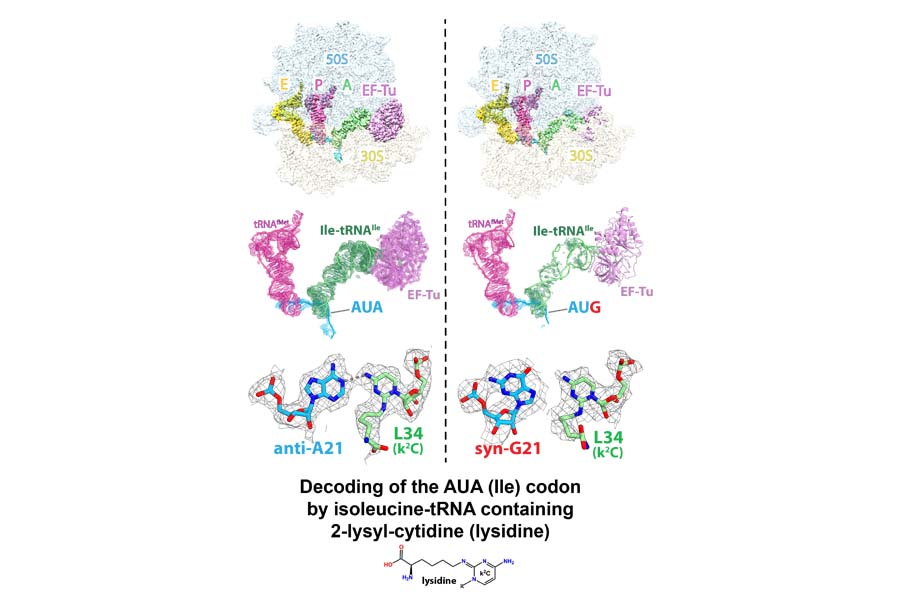
We study how post-transcriptional modifications of RNA improve decoding of the genetic code. The anticodon loop of the minor isoleucine tRNA2Ile in E. coli (C34) is modified to lysidine (L34) by the enzyme TilS. In the absence of modification, the anticodon sequence of Ile-tRNA2Ile is CAU, identical to that of tRNAMet. The modification of C34 in Ile-tRNA2Ile allows accurate decoding of the cognate AUA codon, and avoids the near-cognate AUG codon. We determined the cryo-EM structures of the E. coli ribosome bound to EF-Tu-GDPCP-Ile-tRNAL34Ile and the AUA or AUG codons in the A site. The structures show how lysidine 34 (L34) excludes the near-cognate AUG codon during translation.
-
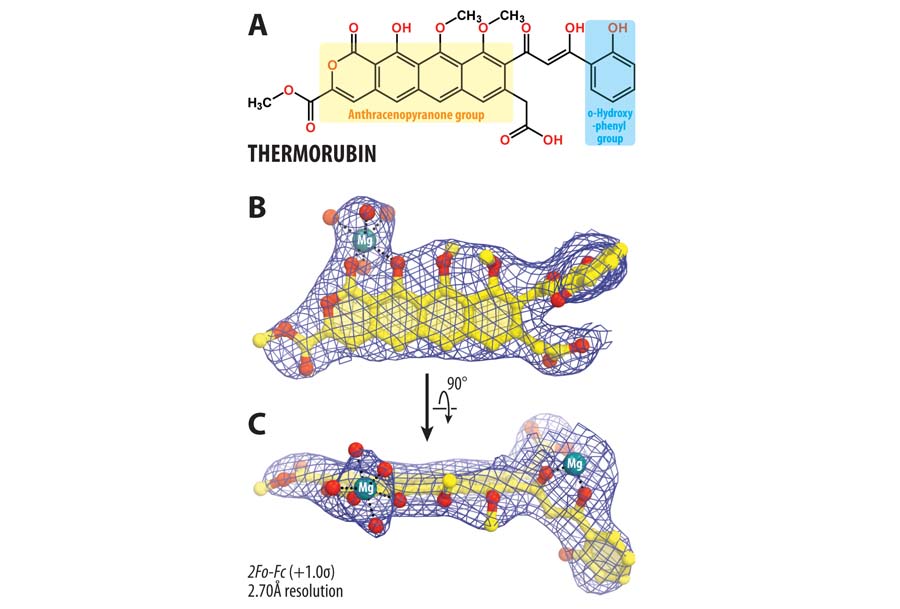
Mechanisms of translation inhibition by ribosome-targeting antibiotics
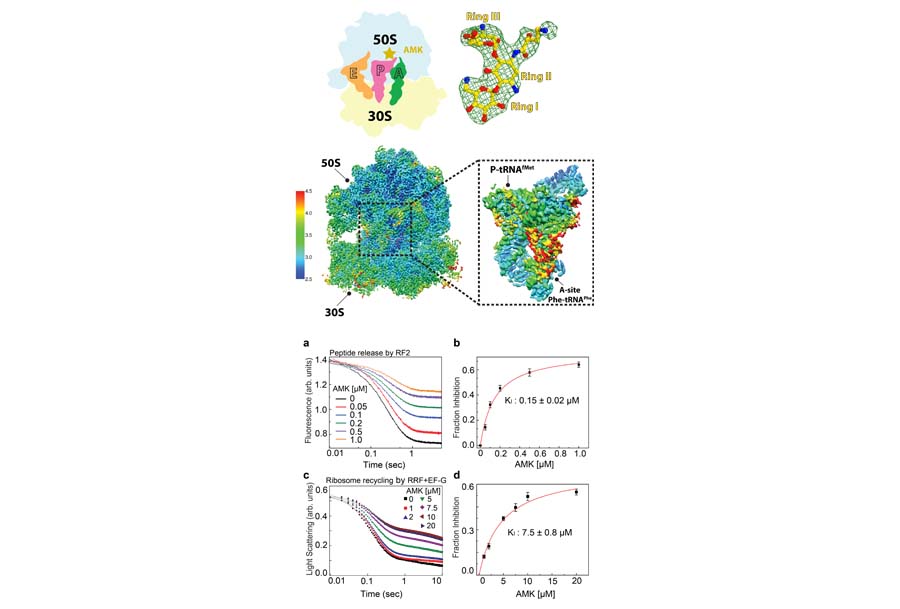
The ribosome is a major target of antibiotics, with more than half of antibiotics used in the clinic targeting the bacterial ribosome. Through national and international collaborations, we recently elucidated the structural bases of inhibition by the antibiotic thermorubin, and the aminoglycoside antibiotics amikacin and kanamycin.
-
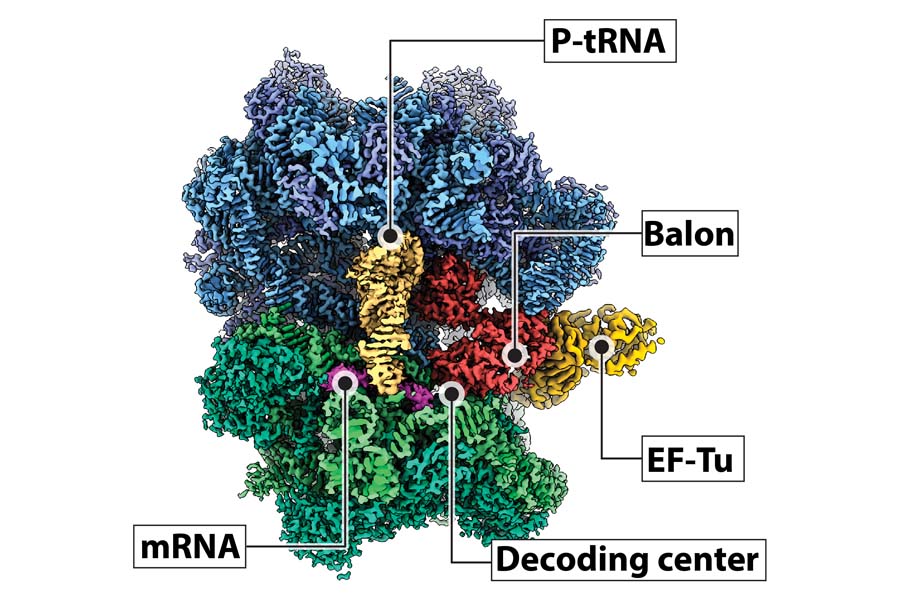
Translational control in Mycobacteria
Mycobacteria use non-canonical and novel strategies to regulate protein synthesis. Mycobacterium tuberculosis, the causative agent of tuberculosis (TB), can undergo extended periods of non-replicating persistence in the host. The underlying mechanisms are not understood; however, translational regulation is thought to play a role. We identified a novel ribosome hibernation factor in M. tuberculosis and M. smegmatis, called Balon, which has a high structural similarity with archaeal and eukaryotic peptide release factors. The peculiarity of this novel ribosome hibernation mechanism is that it can commence while ribosomes are still translating, which likely stems from the slow rates of protein biosynthesis in these bacterial species. Interestingly, Balon-bound ribosomes also associate with the elongation factor-Tu (EF-Tu), revealing an unexpected role for EF-Tu during stress and ribosome hibernation.